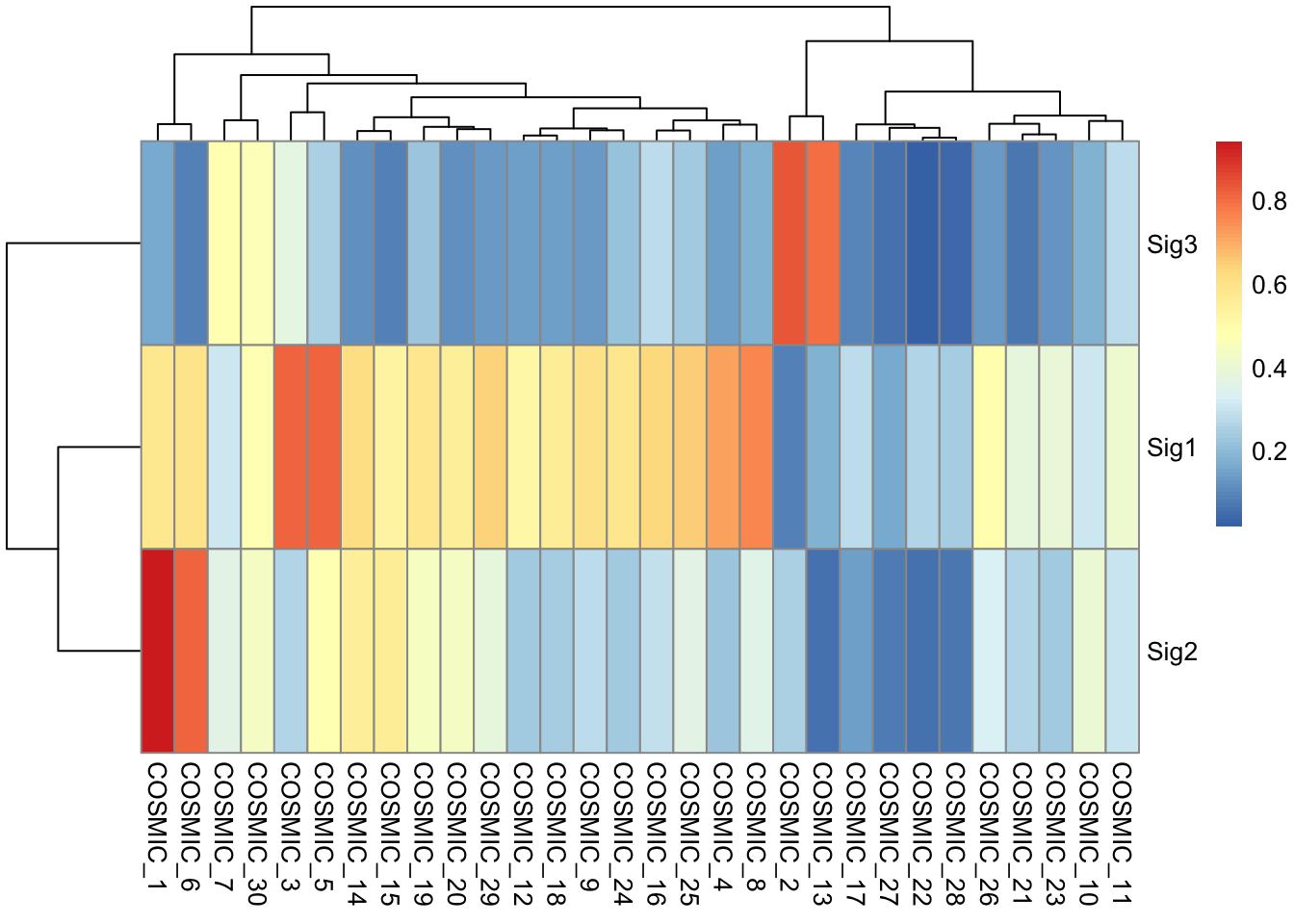
Chapter 2 COSMIC Signature Identification | Sigminer: A Scalable Toolkit to Extract, Analyze and Visualize Mutational Signatures

Reporting workflow for somatic variants. All variants outputs from the... | Download Scientific Diagram

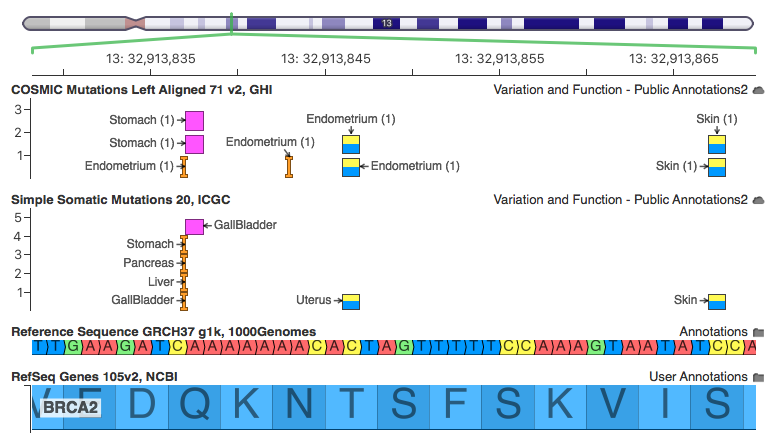
VCF/Plotein: A web application to facilitate the clinical interpretation of genetic and genomic variants from exome sequencing projects | bioRxiv
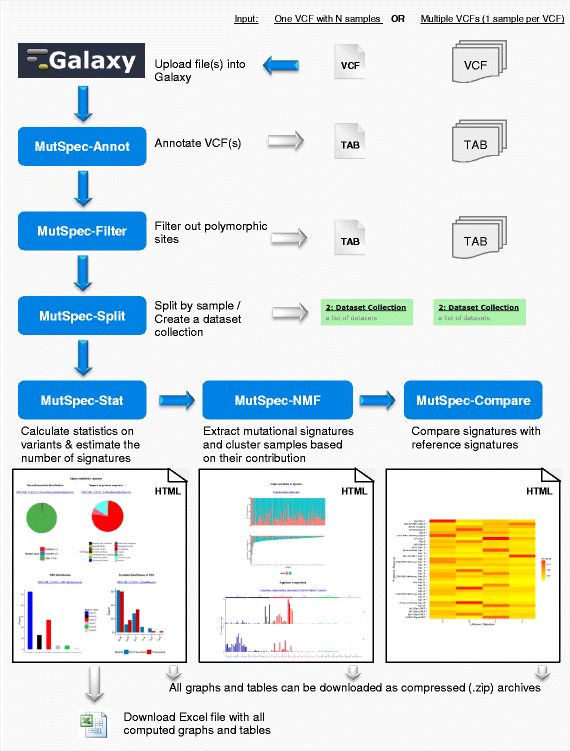
MutSpec: a Galaxy toolbox for streamlined analyses of somatic mutation spectra in human and mouse cancer genomes | BMC Bioinformatics | Full Text
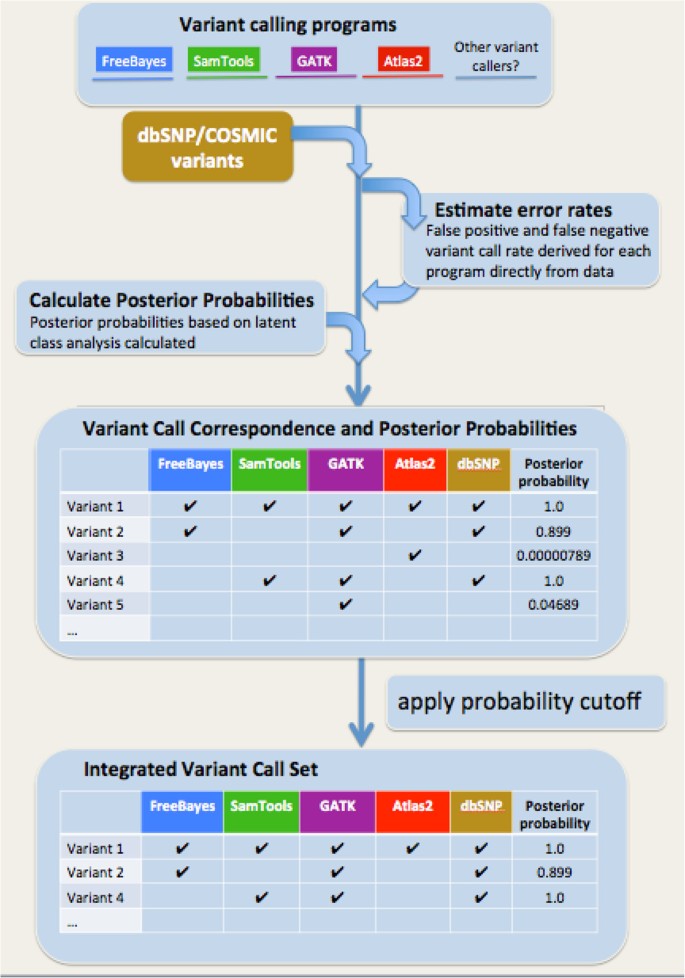
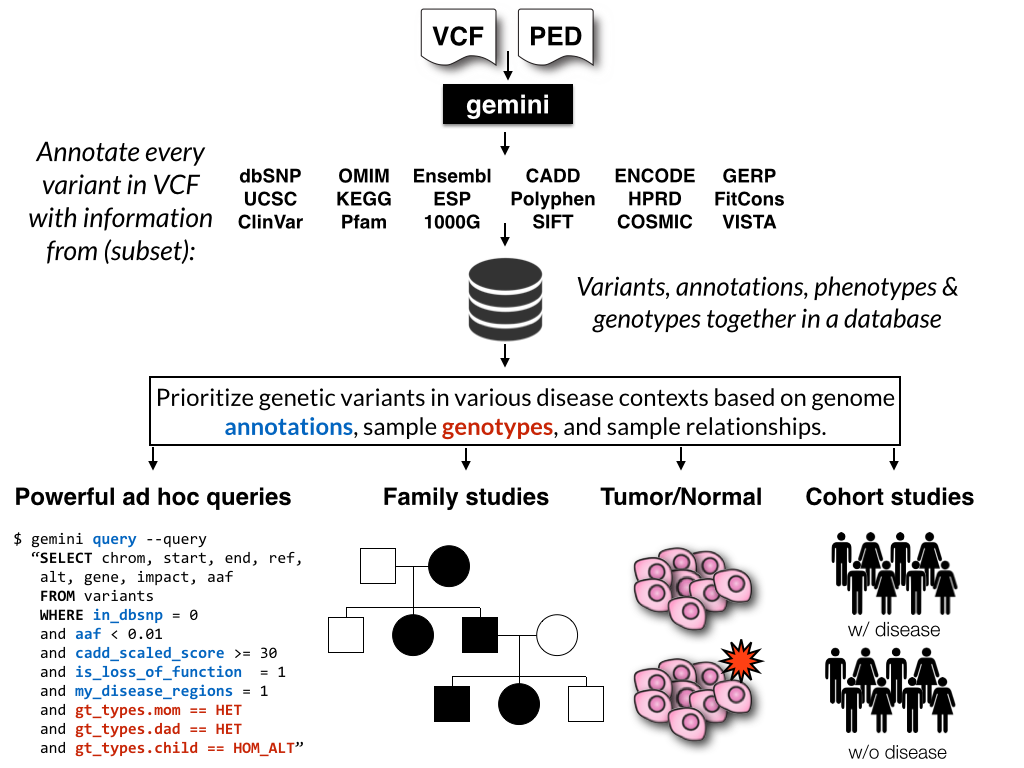
BAYSIC: a Bayesian method for combining sets of genome variants with improved specificity and sensitivity | BMC Bioinformatics | Full Text
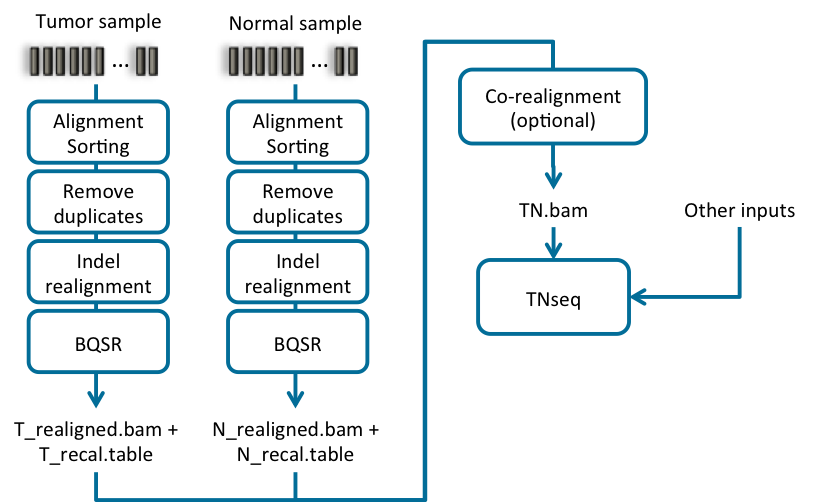
Accelerating Germline and Somatic Genomic Analysis of Whole Genomes and Exomes with NVIDIA Clara Parabricks v.3.6 | by Johnny Israeli | Medium